Research
High throughput molecular connectomics
One of the major obstacles toward understanding how the properties of neurons arise from the structure of their synaptic connections is the lack of tools to probe neuronal connectivity in a scalable manner. Existing approaches, such as serial electron microscopy or multiple patch clamp recordings, are extremely labor-intensive and largely limited to probing local connections. To overcome these limitations, we are working on new ways of using molecular methods to read out neuronal connectivity and relate it to gene expression in individual neurons. Our goal is to apply these new methods to understand the rules that different cell types of cortical neurons follow in selecting their synaptic inputs and how these rules differ between cortical areas and across the mammalian lineage.
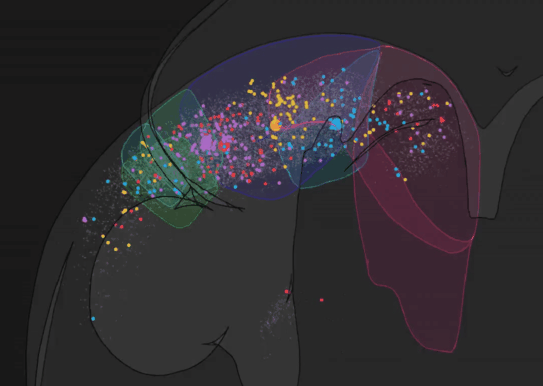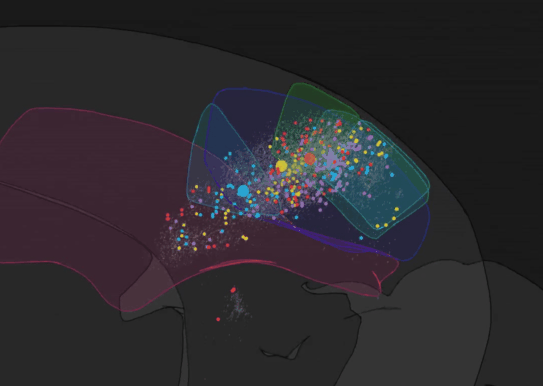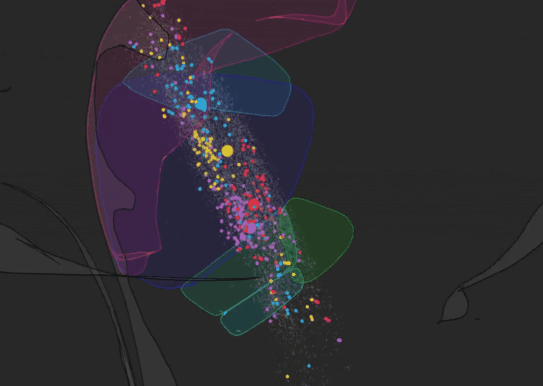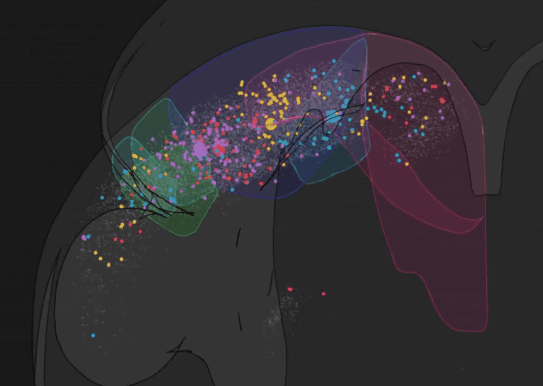
We have recently developed BRISC: Barcoded Rabies In Situ Connectomics, a new circuit tracing method, relying on rabies viruses expressing molecular barcodes, that will make it possible for a single experimenter to rapidly trace thousands of synaptic connections in a matter of weeks. Our approach relies on the ability of rabies virus to spread between synaptically connected neurons. When individual neurons are infected with rabies viruses expressing unique RNA “barcodes”, they transmit their barcodes to their presynaptic inputs. Therefore, synaptic connections can be identified by matching barcode sequences between pre- and post-synaptic neurons. To detect viral barcodes, we use in situ sequencing to read them out from brain sections together with transcripts of cell type markers that allow us to determine the molecular identity of barcoded neurons. We are now applying this method to systematically characterize wiring rules of cortical neurons.

